Welcome to ProkFunFind’s documentation!¶
ProkFunFind is a pipeline that can be used to predict genes related to function of interest. Criteria for function detection include presence of detected genes, and genomic co-localization.
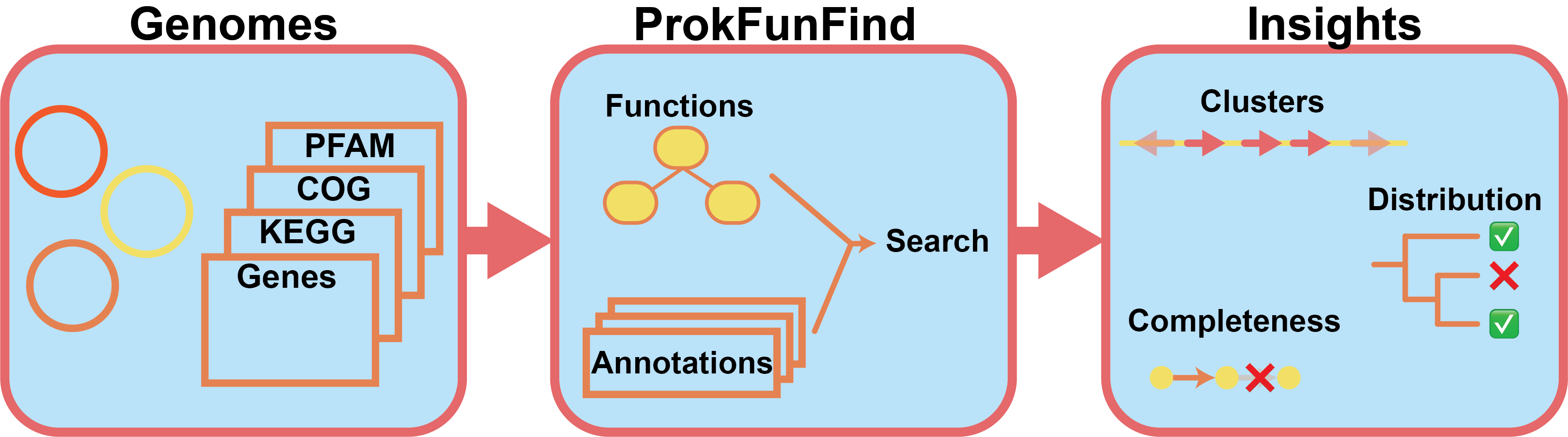
In order to identify genes using ProkFunFind, the user needs:
Select the correct search approaches and filtering parameters to identify your genes of interest
Protein sequences
Profile HMMs
COGs
KOs
Protein domain signatures
Define the gene components for the function system in JSON format
Define the essential and non essential components of the overall function of interest.
Perform the search for the functions in your genomes of interest using ProkFunFind
Analyze the search results
Assess completeness of function in different organisms
Identify potential gene clusters
Analyze species distribution